[NEW PAPER] Receptor-like kinases in green algae: a hidden diversity revealed by modern bioinformatics
Our group investigated the conservation of receptor-like kinases (RLKs) in algae species and recently published a paper on this topic
NEWSPUBLICATIONS
Cell Wall Dynamics Team
1/5/20263 min read
Receptor like kinases (RLKs) are proteins that sit at the cell surface and help cells to detect both external signals and internal changes, thus triggering growth and defense responses essential for survival and adaptation to environmental challenges. These receptors, abundant and well characterized in land plants, are thought to originate from the Chlorophyta division (green algae).
While previous literature has highlighted a limited number of such receptors in green algae species, we speculated that this would be due to technical limitations rather than actual biological reality.
We therefore asked, how could RLKs present in this taxon have been under-examined? Could their structure provide clues on the family’s evolution throughout the green lineage, and how algae perceive their environment?
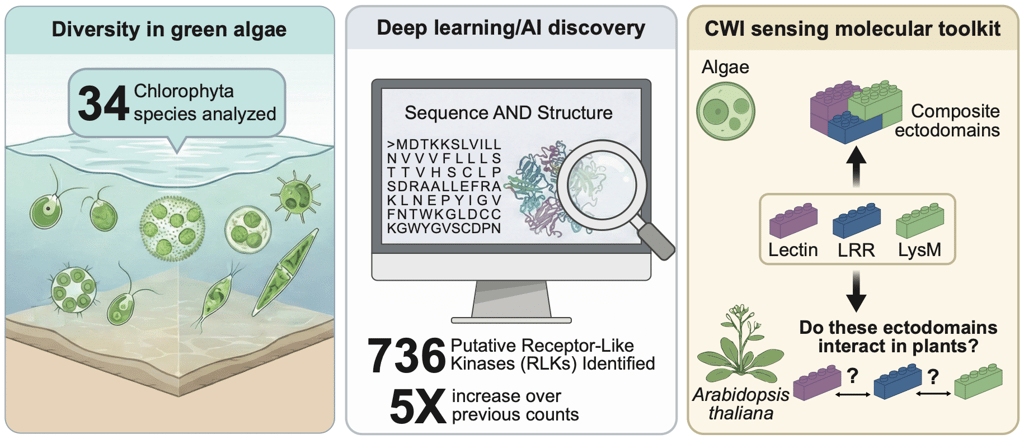
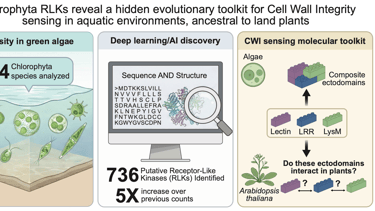
In this context, we combined advanced bioinformatics tools exploiting the latest AI-driven tools to analyze protein data from 34 different Chlorophyta species, looking for protein sequences that matched the structural signatures of RLKs. What we found was striking: 736 putative RLKs, against the 137 previously described, a dramatic expansion of the known repertoire!
When we looked closely at the structures of these proteins, we immediately observed that many have extracellular domains reminiscent of those found in land plant receptors, including: Motifs associated with protein interactions, carbohydrate binding, cell wall remodeling, and even mechanosensing. These features suggest that green algae may have evolved sophisticated mechanisms to monitor their cell wall environment much earlier in evolution than previously appreciated.
Our work not only broadens the catalog of RLKs in Chlorophyta but also establishes an exciting framework for future studies on their roles in cell wall perception and signaling — helping us understand how these systems evolved to the complex receptor networks we observe in terrestrial plants today.
To explore the motivations and technical advances behind this study in more detail, we asked Demetrio and Bastien, co-first authors, a few questions.
1. What motivated you to investigate receptor-like kinases specifically in Chlorophyta?
As always, it came from simple questions. How can algae, which are subjected with a myriads of ever-changing environments, process information from the outside? Why do RLK distribution vary so much from one species to another? Based on recent studies and on our understanding of algal research, we thought the current identification had significant gaps that current technologies could fill.
2. What changed in the software landscape to allow for refined bioinformatic analysis?
The development of deep-learning algorithms has proven incredibly powerful in protein domain identification, mapping, and even structure prediction. At the same time, the quality and sheer number of proteomic data has greatly improved in recent years. Through these advancements, we could significantly improve all levels of our analysis, from family identification to domain annotation.
3. Did you expect this amount and diversity of RLKs?
We would be lying if we said no, but perhaps not to that extent! Very recent evidence indicated that the current number and diversity was very likely underestimated, but our work now brings whole new implications for algae sensing as well as on RLKs’ evolution.
4. How do you see this expanded repertoire of algal RLKs expand in our current understanding of evolution?
Is Chlorophyta truly RLKs’birthplace? Considering the drastic improvements we made over their identification, it is reasonable to hypothesize that RLK could already be present in earlier-diverging species, but we simply lacked the tools to predict them. Moreover, our analysis identified RLKs with complex, multidomain architectures. Such structural complexity may have important implications for receptor–ligand interactions and sensory mechanisms in algae. At a minimum, it suggests that algal sensing pathways may support a higher degree of ligand recognition and response specificity than previously observed.
5. Any last comments?
Please be mindful of the genomic and proteomic resources that you upload: sometimes filtering the usable from the waste can be a huge time sink, especially if dealing with non-model species!
Reference:
Marcianò, D., B. G. Dauphin, F. Basso, C. Funk, and L. Bacete. 2026. “ A Comparative Analysis of Receptor-Like Kinases in Chlorophyta Reveals the Presence of Putative Cell Wall Integrity Sensors.” Physiologia Plantarum 178, no. 1: e70703. https://doi.org/10.1111/ppl.70703.